-Search query
-Search result
Showing 1 - 50 of 367 items for (author: ward & rf)

EMDB-19666: 
Cryo-electron tomogram of apoferritin
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19667: 
Cryo-electron tomogram of keyhole limpet hemocyanin
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19668: 
Cryo-electron tomogram of TGEV virions
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19669: 
Cryo-electron tomogram of Influenza virions
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19670: 
Cryo-electron tomogram of Magnetospirillum gryphiswaldense
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19671: 
In situ cryo-electron tomogram of Saccharomyces cerevisiae
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19672: 
Cryo-electron tomogram of keyhole limpet hemocyanin (stage position)
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19673: 
Cryo-electron tomogram of keyhole limpet hemocyanin (3 um image shift)
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19674: 
Cryo-electron tomogram of keyhole limpet hemocyanin (6 um image shift)
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I
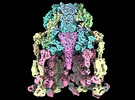
EMDB-29383: 
Structure of baseplate with receptor binding complex of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fqc: 
Structure of baseplate with receptor binding complex of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH
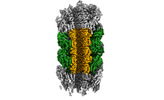
EMDB-29353: 
Structure of Agrobacterium tumefaciens bacteriophage Milano curved tail
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-29354: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-tube
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-29355: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-sheath
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fop: 
Structure of Agrobacterium tumefaciens bacteriophage Milano curved tail
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fou: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-tube
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8foy: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-sheath
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-16453: 
SARS-CoV-2 Omicron Variant Spike Trimer in complex with three 17T2 Fabs
Method: single particle / : Modrego A, Carlero D, Bueno-Carrasco MT, Santiago C, Carolis C, Arranz R, Blanco J, Magri G

EMDB-16473: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)
Method: single particle / : Modrego A, Carlero D, Bueno-Carrasco MT, Santiago C, Carolis C, Arranz R, Blanco J, Magri G

PDB-8c89: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)
Method: single particle / : Modrego A, Carlero D, Bueno-Carrasco MT, Santiago C, Carolis C, Arranz R, Blanco J, Magri G

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

EMDB-29500: 
Portal assembly of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH
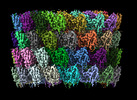
EMDB-29501: 
Collar sheath structure of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-29503: 
Neck structure of Agrobacterium phage Milano, C3 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH
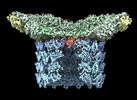
EMDB-29504: 
Structure of neck and portal vertex of Agrobacterium phage Milano, C5 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-29512: 
Structure of tail-neck junction of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-29540: 
Structure of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH
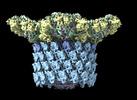
EMDB-29541: 
Structure of neck with portal vertex of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwb: 
Portal assembly of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwc: 
Collar sheath structure of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwe: 
Neck structure of Agrobacterium phage Milano, C3 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwg: 
Structure of neck and portal vertex of Agrobacterium phage Milano, C5 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwm: 
Structure of tail-neck junction of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fxp: 
Structure of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fxr: 
Structure of neck with portal vertex of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-41302: 
Lassa GPC trimer in complex with Fab GP23
Method: single particle / : Gorman J, Kwong PD

EMDB-41298: 
CryoEM structure of Myxococcus xanthus type IV pilus
Method: helical / : Zheng W, Egelman EH

PDB-8tj2: 
CryoEM structure of Myxococcus xanthus type IV pilus
Method: helical / : Zheng W, Egelman EH

EMDB-26859: 
Ligand-free Lassa GPC Trimer with C3 Symmetry
Method: single particle / : Gorman J, Kwong PD

EMDB-14504: 
Three-dimensional structure of myosin binding protein C in rat cardiac muscle
Method: subtomogram averaging / : Luther PK, Morris EP, Huang X, Jun L
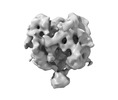
EMDB-26740: 
Ligand-free Lassa GPC Trimer with C1 Symmetry
Method: single particle / : Gorman J, Kwong PD

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
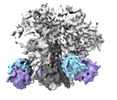
EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
